Environmental pollution by toxic organic compounds is a consequence of industrial development and the exploitation of fossil energy resources. Some pollutants, including polycyclic aromatic hydrocarbons (PAHs), are persistent for long time periods and represent a risk for human health and for ecosystems.
In order to promote bioremediation processes for the treatment of polluted sites, several bacterial species able to degrade PAHs have been isolated and studied by researchers in our laboratory. However, 95% of soil bacteria cannot be cultivated in laboratory and are virtually unknown so that few data are available about species that degrade PAHs
in situ. To identify these bacteria, researchers have implemented molecular methods, independent of laboratory culture.
 | 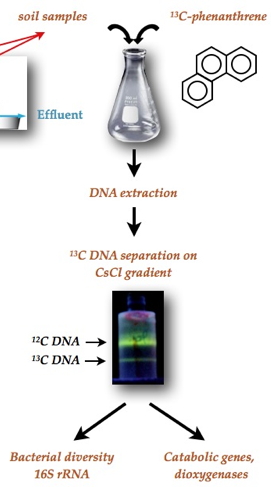 |
|
The approach used is based on in situ metabolic labeling of target bacteria able to use PAHs as carbon sources. To this end, 13C labeled phenanthrene was synthesized and introduced as a substrate in samples of contaminated soil. After incubation in microcosms, 13C-labeled and unlabeled DNA was extracted from soil, and separated. The 13C-enriched DNA fraction from bacteria having metabolized phenanthrene was then used to identify the bacteria, showing that the main actors of degradation were ß-proteobacteria, mostly belonging to non-cultivable genera [1]. The study also revealed that the nature of the dominant bacteria changed during the incubation, suggesting rapid adaptation of the microbial community to the bioavailability of PAHs in soil. |
To explore the biocatalytic potential of the identified species, the catalytic domain of dioxygenase genes (enzymes that catalyze the initial oxidation step of PAHs) was amplified from the
13C-DNA extracted from soil. The majority (82%) had little in common with known enzymes. The catalytic activity associated with these sequences was then investigated by constructing hybrid enzymes
[2] but most of them proved unstable. In order to reconstruct the genome of the bacteria most active in PAH degradation, a
metagenomics approach was undertaken in collaboration with the Genoscope at Evry. Of the 34 000 predicted protein sequences, 2/3 appeared to be related to those of ß-proteobacteria, among which are the dominant genera identified above.
This type of study shed light on bacteria that play a major role in the in situ biodegradation of PAHs and might provide tools to improve processes for bioremediation of polluted sites.
Microcosm: device reproducing at small scale a complex ecosystem.
The metagenomics approach allows to analyze the genomes of a mixed population of bacteria without prior isolation of its constituent species.