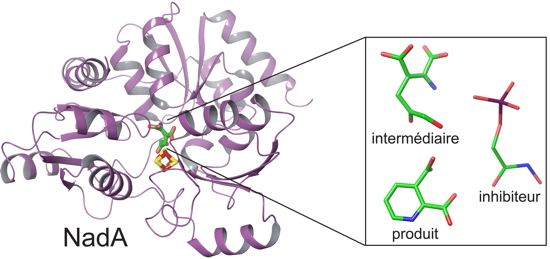
The enzyme NadA is of great interest to scientists as it is involved in the synthesis of an essential cofactor in the metabolism of pathogenic organisms, such as Helicobacter pylori and Microbacterium leprae. While the former is a bacterium related to ulcer and gastric cancer, the latter is responsible for leprosy. Using a crystallography technique in a controlled atmosphere, researchers from CEA-IBS and from our laboratory have deciphered the catalytic mechanism of this enzyme in anaerobic conditions (i.e., without oxygen) necessary for its operation and close to the enzymes found in the stomach.
"
We observed an aggregate consisting of four iron atoms and four sulphur atoms, where a catalytic reaction occurs that is essential for the enzyme to function", says Juan C. Fontecilla-Camps, researcher at CEA-IBS. With this in mind, the scientists produced two mutations of the catalytic site: one to stop the reaction catalysis at an intermediate stage (which is normally unstable), and the other to trap the product of the reaction. "
We were able to monitor the reaction from a structural point of view by analyzing both the intermediate and the product", the scientist says. "
What we observed is that the product is redirected towards the catalytic center of the protein. This result is quite unexpected for this enzyme." The researchers are now seeking to explore the possibilities of finding inhibitory molecules capable of binding to the different conformations that were identified. These inhibitors blocking the function of the enzyme NadA could then become suitable for the design of new antibiotics.