Nitrous oxide (N
2O) is an ozone-depleting agent but also a powerful greenhouse gas with a warming potential 300 times greater than that of CO
2
[1]. Its main sources are the nitric and adipic acid production industries, intensive agriculture and road traffic. N
2O also has direct harmful effects on health since it can cause, under conditions of prolonged exposure, or severe brain disorders and even irreversible DNA damage. Its ability to inhibit methionine synthase has been connected to these pathologies
[2, 3]. In the latter case, the development of rapid and reliable detection systems is therefore of obvious interest. Current methodologies, based on Fourier transform infrared or gas chromatography, are very expensive to set up and ultimately not very accurate.
In order to apprehend the electronic and structural parameters required to master N
2O activation, we decided to get inspired from Nature since N
2O is also a substrate used by certain organisms. The co-called copper-containing nitrous oxide reductase (N
2Or) is indeed capable of the two-electron reduction of N
2O, this reaction occurring during the nitrification/denitrification cycles of bacteria. The X-ray crystal structure shows the presence at the active site of a unique tetranuclear copper cluster with a N/S environment
[4-6]. The mechanism has been dissected for more than a decade by combining experimental and sophisticated theoretical approaches. All the evidences gathered indicate the presence of a key intermediate (Figure 1). N
2O would be coordinated by two of the four metal centers and the overall structure stabilized by a hydrogen bond
[7].
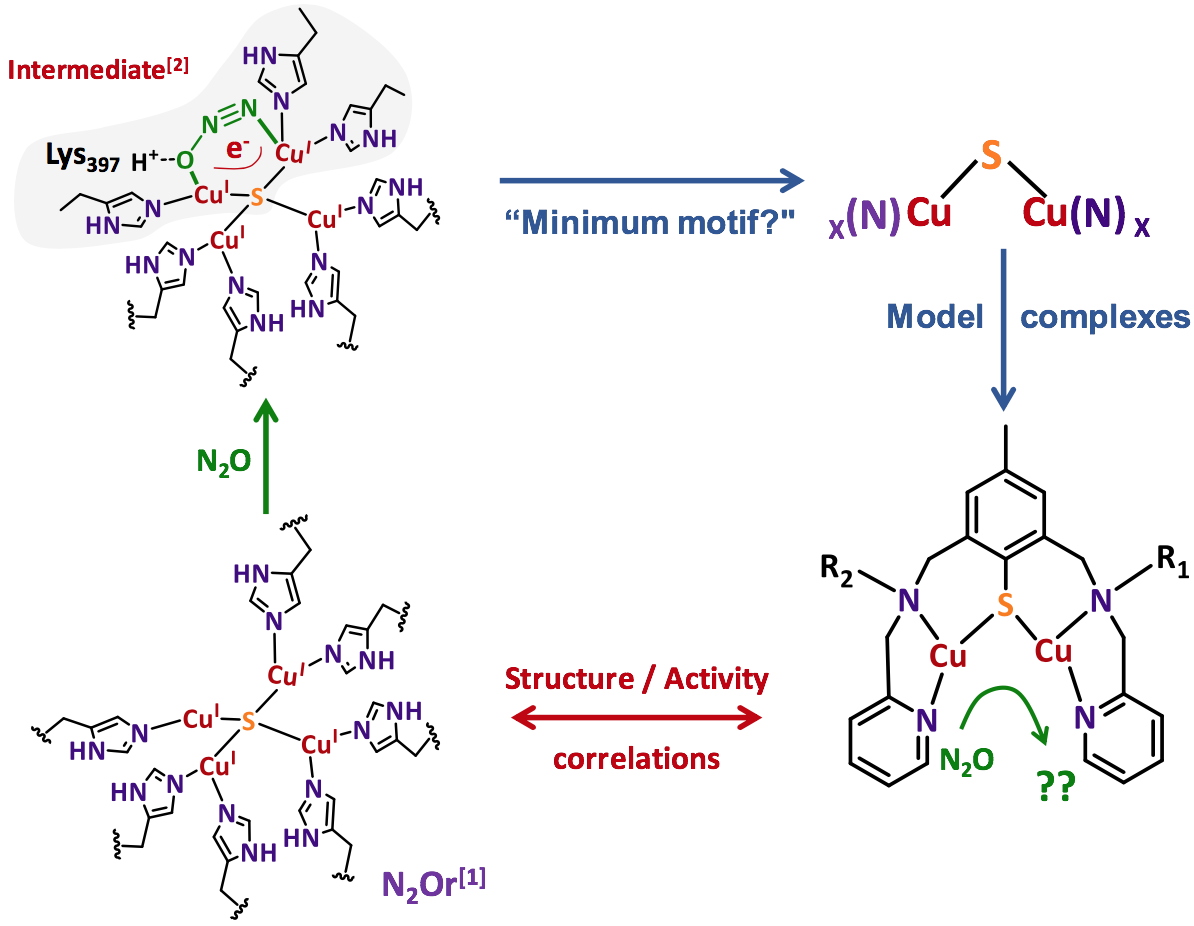 Figure 1
Figure 1. N
2Or active site, proposed intermediate and bio-inspired approach for the preparation of dinuclear copper complexes with N/S environment.
Coord. Chem. Rev.
2019, 387, 436-449
Chem. Rev.
2014, 114, 3689-3853
J. Am. Chem. Soc.
2017, 139(12), 4462-4476
In our approach, the hypothesis was to restrict the active site to its minimum motif required for the activity. Given the bi-electronic nature of the reduction process and the structure of the proposed intermediate, the use of dinuclear copper complexes with a N/S environment was privileged. From the general representation shown in Figure 1, a series of dinuclear species was prepared and characterized prior to reactivity studies. The objective is to be capable of (i) extracting a rationale from structure/activity correlations and (ii) proposing more and more sophisticated and efficient compounds.
The Cu2NxS (x = 2 or 3) motifs isolated from the disulfide ligand precursors are depicted in Figure 2. Each Cu coordination sphere is unique and corresponds to the structural diversity targeted for further reactivity studies.
All the complexes are in a mixed-valent (MV) state, CuIICuI and their electronic properties were examined by combining experimental technics such as Electron Paramagnetic Resonance (EPR), UV-vis/NIR spectrophotometry, Nuclear Magnetic Resonance (NMR) electrochemistry and X-ray diffraction with theoretical methods (DFT and TDDFT, collaboration with Dr. M. Orio – iSm
2 Marseille, France). The delocalized MV states have complicated EPR signatures and the corresponding parameters can only be determined by simulation whereas for localized species, typical spectra of mononuclear Cu
II centers are obtained.
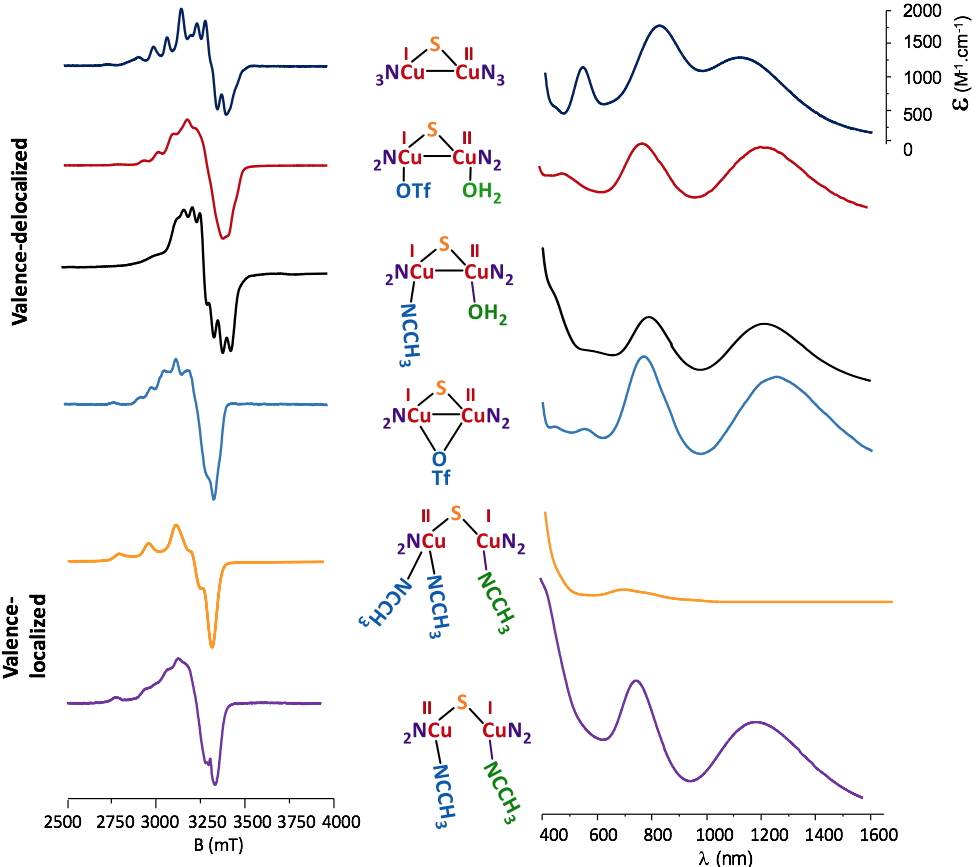 Figure 2
Figure 2. Characteristic EPR and UV-vis/NIR spectra for the isolated copper complexes.
The UV-vis/NIR spectra are also indicative of the delocalized valence with signatures in the near-infrared region (1200-1400 nm). The electrochemical properties are also structure-dependent and sensitive (as for EPR and UV-vis/NIR) to the nature of the solvent, indicating changes within the molecular structure in solution. Finally, X-ray studies (Figure 3) allowed a characterization in the solid state that was combined with DFT and TDDFT methods to determine the exact nature of a given complex upon solvation in acetonitrile (MeCN) and acetone (Me
2CO) used for reactivity studies with N
2O.
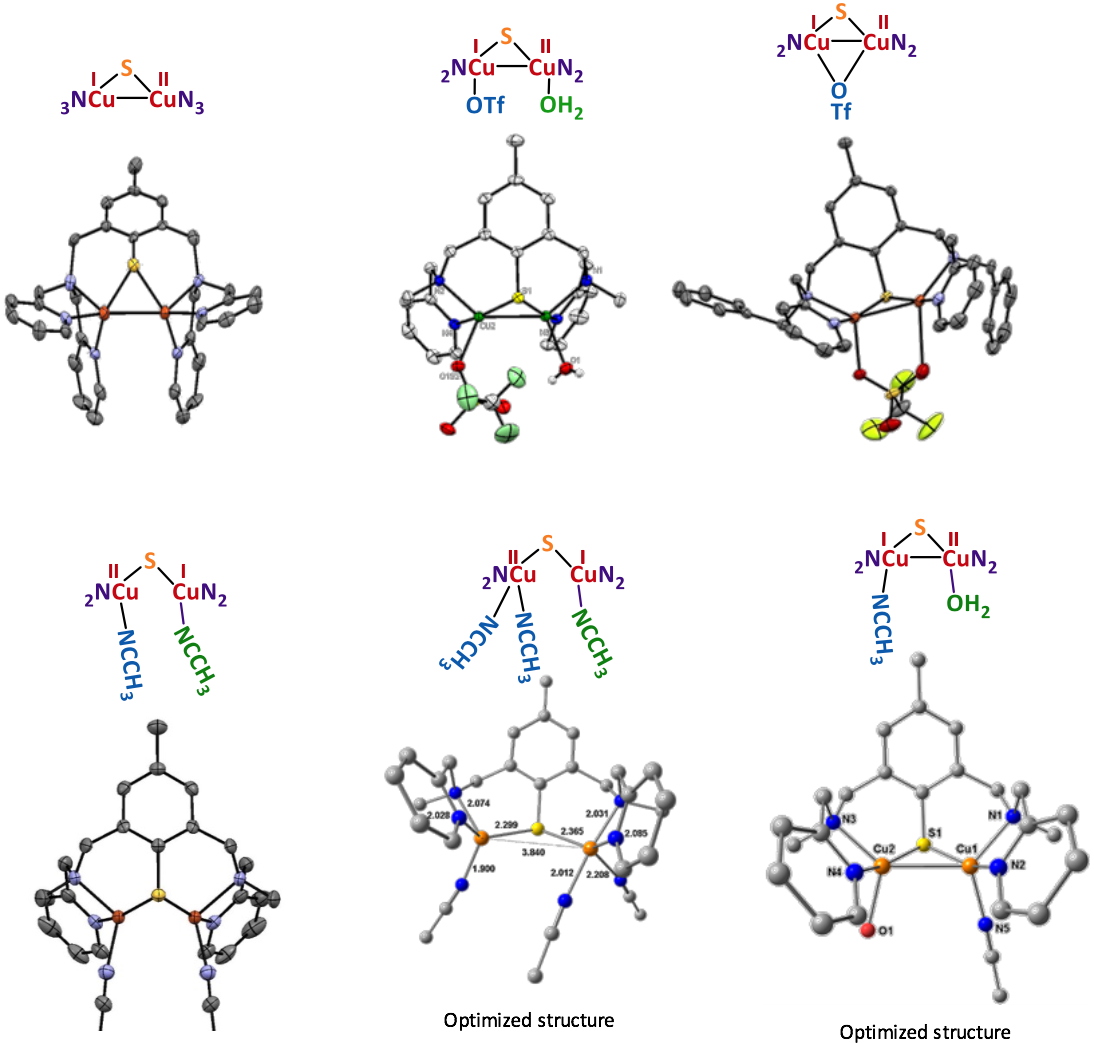 Figure 3
Figure 3. X-ray or optimized structures for the isolated MV complexes.
With the knowledge of the solution structures in hands, stoichiometric N2O reduction experiments were performed. The main result is that only the complexes having a coordinated water molecule from start are active (Figure 4).
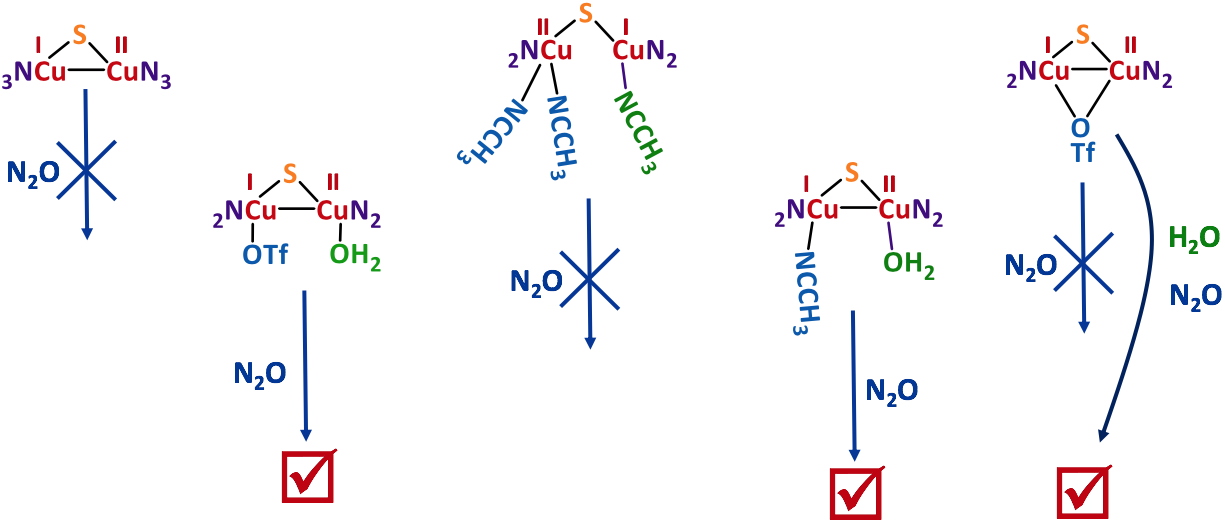 Figure 4
Figure 4. Stoichiometric N
2O reduction by several isolated MV Cu complexes.
The labile character of this weak ligand appears thus to be necessary for N
2O coordination. As an example, a mechanistic study with an active complex is shown in Figure 5.
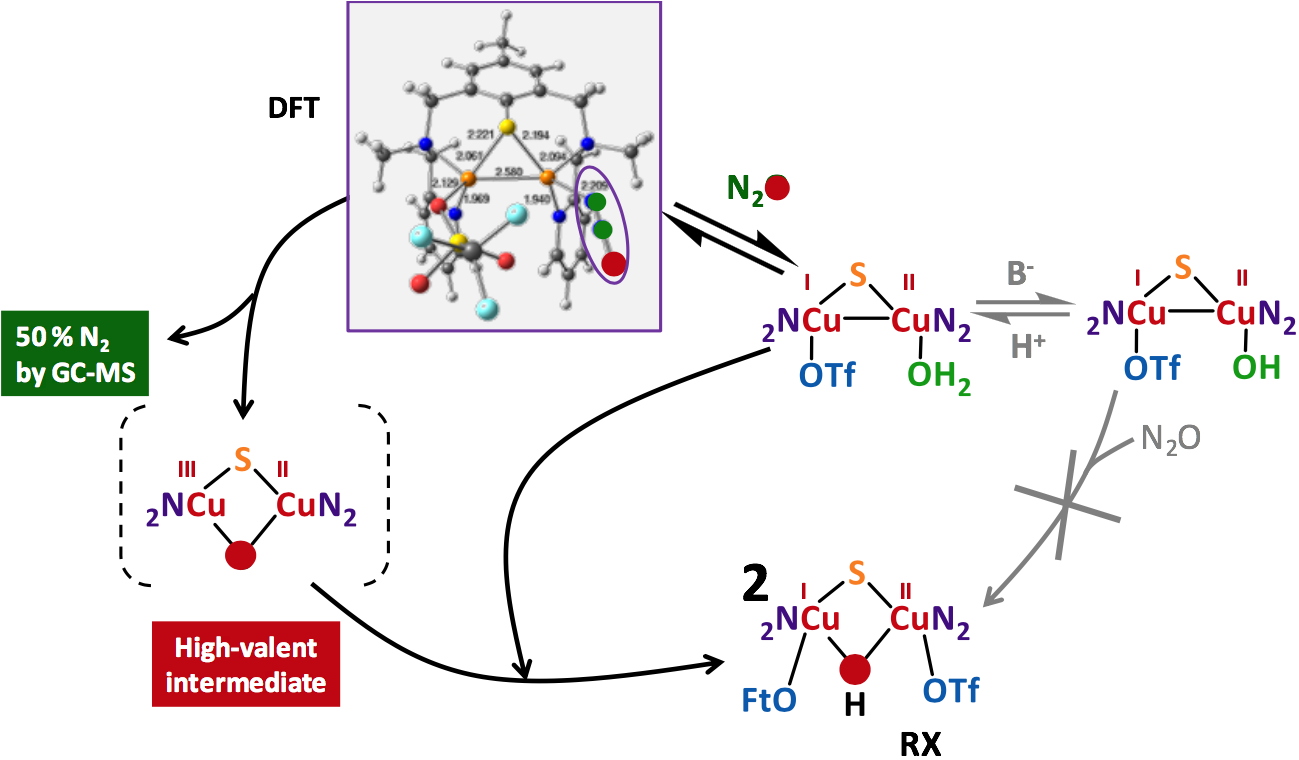 Figure 5
Figure 5. Proposed mechanism for the N
2Or activity of a copper complex having a coordinated water molecule.
The active complexes remain the only species of that kind reported in the literature for mild and stoichiometric N2O activation.
These results were published in the following articles:
• Esmieu C, Orio M, Ménage S, Torelli S.
Inorg. Chem.
2019, 58 (17), 11649-11655
• Esmieu C, Orio M, Mangue J, Pécaut J, Ménage S, Torelli S.
Chem. Eur. J.
2018, 24 (20), 5060-5063
• Mangue J, Dubreucq Q, Pécaut J, Ménage S, Torelli S.
ChemistrySelect
2016, 1 (20), 6345-6348
• Esmieu C, Orio M, Le Pape L, Lebrun C, Pécaut, J, Ménage S, Torelli S.
Inorg. Chem.
2016, 55 (12), 6208-6217
• Esmieu C, Orio M, Torelli S, Le Pape L, Pecaut J, Lebrun C, Ménage S.
Chem. Sci.
2014, 5 (12), 4774-4784
• Torelli S, Orio M, Pécaut J, Jamet H, Le Pape L, Ménage S.
Angew. Chem. Int. Ed.
2010, 49 (44), 8249-8252
References
[1] Ravishankara, A. R.; Daniel, J. S.; Portmann, R. W., Science
2009, 326 (5949), 123-125
[2] Weimann, J., Best Practice & Research Clinical Anaesthesiology
2003, 17 (1), 47-61
[3] Deacon, R.; Lumb, M.; Perry, J.; Chanarin, I.; Minty, B.; Halsey, M.; Nunn, J., Eur. J. Biochem.
1980, 104 (2), 419-422
[4] Pauleta, S. R.; Dell'Acqua, S.; Moura, I., Coord. Chem. Rev.
2013, 257 (2), 332-349
[5] Dell’Acqua, S.; Pauleta, S.; Paes de Sousa, P.; Monzani, E.; Casella, L.; Moura, J.; Moura, I., J. Biol. Inorg. Chem.
2010, 15 (6), 967-976
[6] Pomowski, A.; Zumft, W. G.; Kroneck, P. M. H.; Einsle, O., Nature
2011, 477 (7363), 234-237
[7] Pauleta, S. R.; Carepo, M. S. P.; Moura, I., Coord. Chem. Rev.
2019, 387, 436-449