The fight against the resistance of pathogenic bacteria to antibiotics is one of the major health challenges. A new approach to anti-virulence is to target the biological systems that bacteria put in place to meet their essential nutrient needs. The Ferric Uptake Regulator (FUR) is a global transcriptional regulator that controls the expression of genes involved in iron homeostasis, virulence and oxidative stress. Ubiquitous in Gram-negative bacteria but absent from eukaryotes, FUR is a prime target since the inactivation of its gene in various pathogens attenuates their virulence.
In collaboration with the Department of Molecular Chemistry (University of Grenoble Alpes) and the Institute of Structural Biology of Grenoble, researchers from our laboratory develop FUR inhibitors. To achieve such inhibitors, they first screened a library of combinatorial chemistry of peptide aptamers for their ability to inhibit binding of FUR to DNA. These aptamers are constituted by a variable peptide loop of 13 amino acids constituting the active part anchored to a constant protein base. Secondly, only the active loops were studied in the form of linear peptides of 8 to 13 amino acids in order to develop smaller anti-FUR molecules
[1]. The researchers then identified the essential residues involved in the inhibition and in the inhibitor peptide / target protein interaction zone.
In recent works
[2], they studied the inhibitory properties of the most active peptides previously identified in order to characterize their interaction with FUR and to understand the involved mechanisms of inhibition. The researchers were able to identify two types of peptides that either inhibit the dimerization of FUR or its binding to DNA (
Figure). With the combination of mutagenesis, inhibition tests, double-hybrid, ITC (IsoThermal Calorimetry) and molecular modeling, they identified the residues involved in the binding pocket of FUR to DNA. They showed that the bound peptides sterically hinder the accessibility of DNA to its binding site on FUR, effectively inhibiting the protein
[3].
This work, which targets a system as essential as the regulation of iron homeostasis in bacteria, has allowed the development of small peptides with anti-bacterial action. It will be interesting to test the specificity of these inhibitions on FUR proteins of other pathogens as well as on a class of newly identified tetrameric FURs
[4].
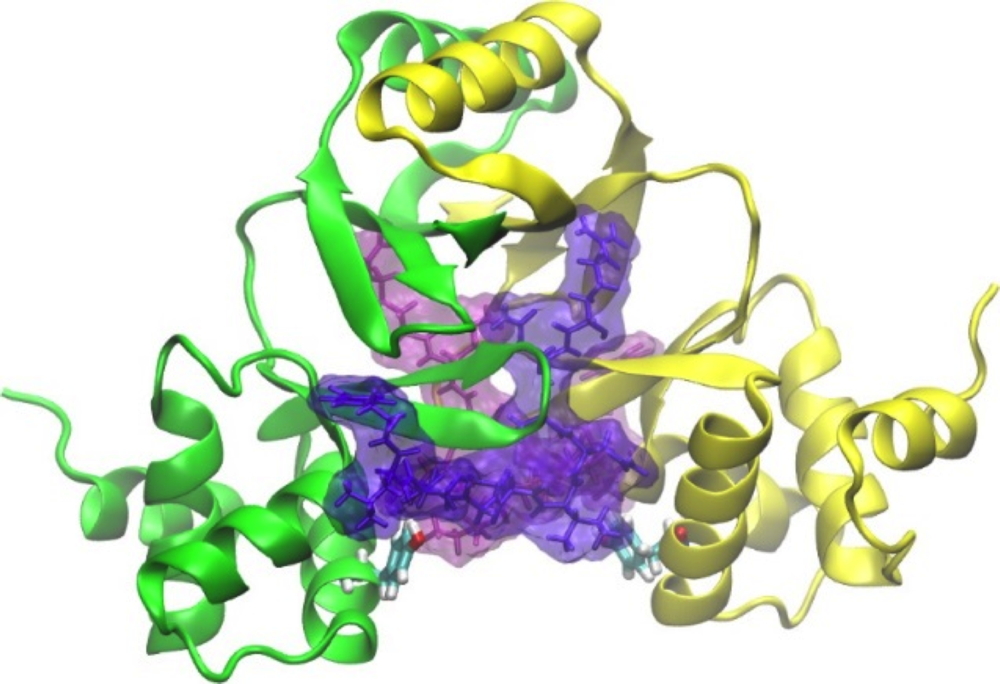
A structure model of 2 inhibitory peptides (pink and violet) simultaneously moored at symmetrical sites on the Escherichia coli FUR protein and resulting from the coupling between inhibition experiments and modeling. The researchers used "docking" experiments to illustrate that these two peptides can bind simultaneously to distinct sites on the FUR dimer, one per subunit.
Yellow: monomer A of FUR; Green: monomer B of FUR.
These research were supported by an ANR, the Arcane and GRAL LabEx as well as by the CEA.