The discovery of penicillin a hundred years ago was a major medical breakthrough that initiated the development of antibiotics. However, their intensive or unsuitable use led to the emergence of pathogens resistant or multi-resistant to all classes of antibiotics. This dramatic situation is leading scientists to search for (i) new molecules whose structure and mode of action differ and (ii) new targets in order to prevent resistance.
In the BioCat team, we are concerned about this problem of bacterial multidrug resistance (MDR) and two projects emerged in this context. One project is related to
NAD biosynthesis in bacteria as a promising target pathway; the second project is focusing on
ribosomally synthesized and post-translationally modified peptides (RiPPs) as new molecules against resistant and MDR strains.
Biosynthesis of Nicotinamide Adenine Dinucleotide
S. Ollagnier de Choudens
Nicotinamide adenine dinucleotide (NAD) biosynthesis pathway constitutes an interesting metabolic pathway for the discovery of new antibacterial agents. Its biosynthesis involves the formation of quinolinic acid (QA), the biosynthesis of which is different in prokaryotes and eukaryotes. In eukaryotic organisms, QA is produced by the degradation of L-tryptophan, whereas in most prokaryotes, QA is generated from L-aspartate through the concerted action of two enzymes, the L-aspartate oxidase (NadB) and the quinolinate synthase (NadA), a 4Fe-4S containing enzyme (Figures 1 and 2). Besides these de novo syntheses of NAD, a salvage pathway exists in some organisms that enable NAD to be recycled (Figure 1). However, some pathogens such as
Mycobacterium leprae and
Helicobacter pylori were reported to lack this salvage pathway. The presence in prokaryotes and eukaryotes of distinct pathways for QA biosynthesis, along with the absence of the salvage pathway in some pathogenic microorganisms, makes NadA a target for the development of antibacterial agents. NadA, is a [4Fe-4S]-containing enzyme, that catalyzes the condensation between iminoaspartate (IA), generated by NadB from L-asparatate, and dihydroxyacetone phosphate (DHAP) forming quinolinic acid (QA). This process produces two water molecules and one molecule of inorganic phosphate. During the last years, we progressed extensively in the mechanism of the reaction catalysed by NadA using structural and biochemical approaches and we identified new specific inhibitors based on the first discovered inhibitor (see publications). Concerning mechanistic aspects, in the next years we aim at: (i) trapping substrates in the two active site cavities; (ii) understanding the condensation mechanism, (iii) unravelling the stereochemistry of the reaction. Concerning the antibacterial aspect, we aim at (i) optimizing the known DTHAP-derivatives inhibitors that target the Fe-S cluster; (ii) designing new inhibitors (that do not target the Fe-S cluster) and (iii) investigating QA biogenesis pathway in Gram (+) bacteria, which use different substrates (acetyl-CoA and formate) than Gram (-) bacteria and likely a different mechanism.
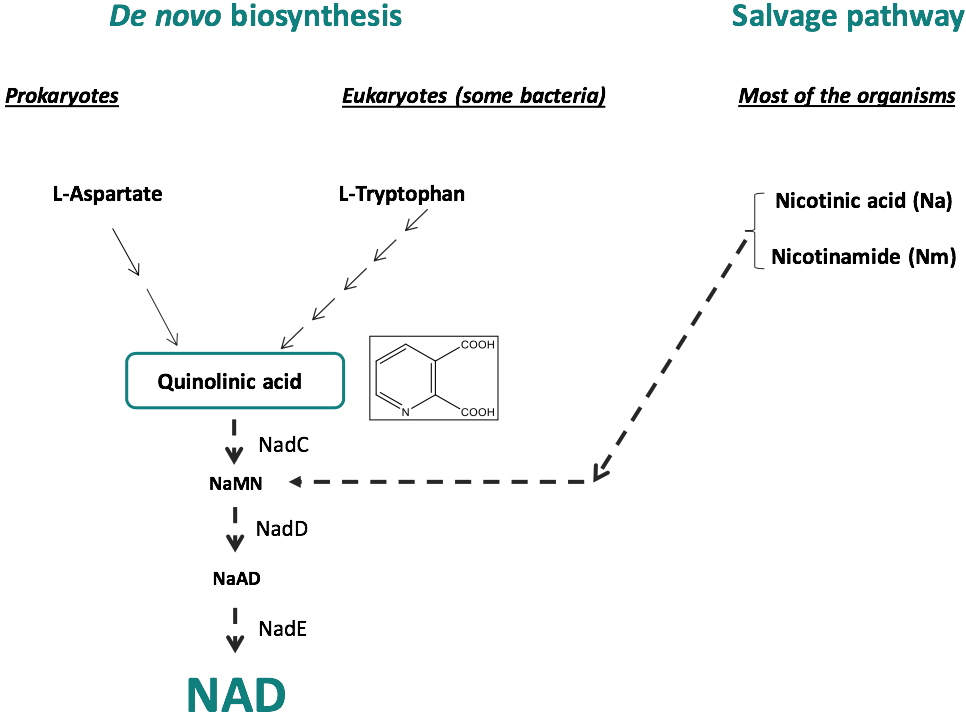
Figure 1: NAD biosynthesis:
De novo biosynthesis and Salvage pathway.
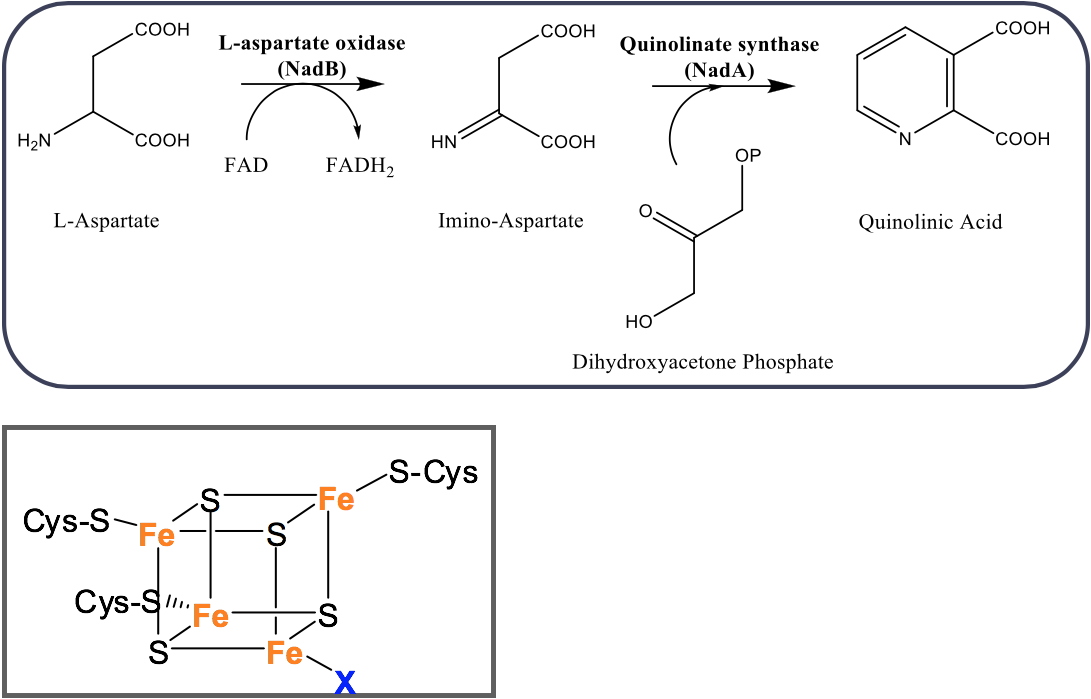
Figure 2: (A) Formation of QA in most of prokaryotes using NadB and NadA enzymes. (B): Fe-S cluster of NadA.
 Collaborations
Collaborations
J.C. Fontecilla-Camps (Métalloprotéines, IBS, Grenoble); P. Amara (Métalloprotéines, IBS, Grenoble); H. de Reuse and S. Cole (Institut Pasteur, Paris); O. Hamelin (BioCE, LCBM); M.O. Fauvarque (BGE, CEA-Grenoble); Y. Sing Wong (DPM, UGA, Grenoble).
 Fundings
Fundings
National Agency of Research (ANR NADBiO, ANR NADIN), GRAL Labex, ARCANE Labex.
 PublicationsSaez Cabodevilla J, Volbeda A, Hamelin O, Latour JM, Gigarel O, Clémancey M, Darnault C, Reichmann D, Amara P, Fontecilla-Camps JC and Ollagnier de Choudens S
PublicationsSaez Cabodevilla J, Volbeda A, Hamelin O, Latour JM, Gigarel O, Clémancey M, Darnault C, Reichmann D, Amara P, Fontecilla-Camps JC and Ollagnier de Choudens S
Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chemical Communications, 2019,
55(26): 3725-3728
Volbeda A, Saez Cabodevilla J, Darnault C, Gigarel O, Han TH, Renoux O, Hamelin O, Ollagnier-de-Choudens S, Amara P and Fontecilla-Camps JC
Crystallographic trapping of reaction intermediates in quinolinic acid synthesis by NadA.
ACS Chemical Biology, 2018,
13(5): 1209-1217
Cherrier MV, Ollagnier de Choudens S and Fontecilla-Camps JC Fe
4S
4 Quinolinate Synthase (NadA).
Encyclopedia of Inorganic and Bioinorganic Chemistry, 2016
Volbeda A, Darnault C, Renoux O, Reichmann D, Amara P, Ollagnier de Choudens S and Fontecilla-Camps JC Crystal structures of quinolinate synthase in complex with a substrate analogue, the condensation intermediate, and substrate-derived product.
Journal of the American Chemical Society, 2016,
138(36): 11802-11809
Reichmann D, Couté Y and Ollagnier De Choudens S Dual activity of quinolinate synthase: Triose phosphate isomerase and dehydration activities play together to form quinolinate.
Biochemistry, 2015,
54(42): 6443-6446
Cherrier MV, Chan A, Darnault C, Reichmann D, Amara P, Ollagnier de Choudens S and Fontecilla-Camps JC The crystal structure of Fe
4S
4 Quinolinate synthase unravels an enzymatic dehydration mechanism that uses Tyrosine and a hydrolase-type triad.
Journal of the American Chemical Society, 2014,
136(14): 5253-5256
Chan A, Clémancey M, Mouesca JM, Amara P, Hamelin O, Latour JM and Ollagnier de Choudens S Studies of inhibitor binding to the [4Fe-4S] cluster of quinolinate synthase.
Angewandte Chemie International Edition, 2012,
51(31): 7711-7714
Rousset C, Fontecave M and Ollagnier de Choudens S The [4Fe-4S] cluster of quinolinate synthase from
Escherichia coli: Investigation of cluster ligands.
FEBS Letters, 2008,
582(19): 2937-2944
Murthy UMN, Ollagnier-de-Choudens S, Sanakis Y, Abdel-Ghany SE, Rousset C, Ye H, Fontecave M, Pilon-Smits EA and Pilon M Characterization of
Arabidopsis thaliana SuFE2 and SuFE3: Functions in chloroplast iron-sulfur cluster assembly and NAD synthesis.
Journal of Biological Chemistry, 2007,
282(25): 18254-18264
Ollagnier-de Choudens S, Loiseau L, Sanakis Y, Barras F and Fontecave M Quinolinate synthetase, an iron-sulfur enzyme in NAD biosynthesis.
FEBS Letters, 2005,
579(17): 3737-3743
Biosynthesis of Ribosomally synthesized and Post-translationally modified Peptides (RiPPs)
V. Duarte, M. Atta, C. Basset, L. Shamseddine (PhD CFR CEA 2020-2023)
Biosynthesis of RiPPs involve diverse enzyme families. Among them,
the radical S-Adenosylmethionine (SAM) enzymes to which we are interested in. Our work focuses on thioether bond containing antimicrobial peptides, namely sactipeptides (sulfur to alpha carbon thioether peptides) and ranthipeptides (radical non-α thioether peptides). In the sactipeptide subclass, radical SAM enzymes introduce intramolecular thioether cross-links between cysteine sulfur and the α-carbon of a partner amino acid. However, in the case of the ranthipepdes, the thioether bound involves either the ß- or the γ- carbon of the partner residue (Figure 3).

Figure 3: Biosynthesis of sactipeptides and ranthipeptides by Radical-SAM.
Our research work investigates novel radical SAM enzymes involved in the biosynthesis of thioether bond containing antimicrobial peptides. Besides the
basic interest to these enzymes, we are interested by the RiPPs biosynthesis from a health point of view, as RiPPs constitute promising antimicrobial agents.
Numerous studies show that the intestinal microflora, well-known as intestinal microbiome, plays a very important role in the health of the host. Bacteriocin production is one of the major protective mechanisms implemented by commensal bacteria. We recently initiated a collaborative project on RumC antimicrobial peptides produced by
Ruminococus gnavus E1, a symbiont present in the digestive tract of healthy adults. Our team developed an expression system in
Escherichia coli to generate a mature RumC1 peptide strictly identical to the one naturally produced and purified from the caecal contents of
R. gnavus E1 mono-associated rats. This
in vitro production of RumC1 was a crucial point, as it was required to produce the mature peptide in suitable amounts for biochemical and structural characterizations as well as antimicrobial assays. We demonstrated that RumC1 belongs to the sactipeptide group of RiPPs. Combined site directed mutagenesis experiments and mass spectrometry analyses led to identify the post-translational modifications of RumC1.
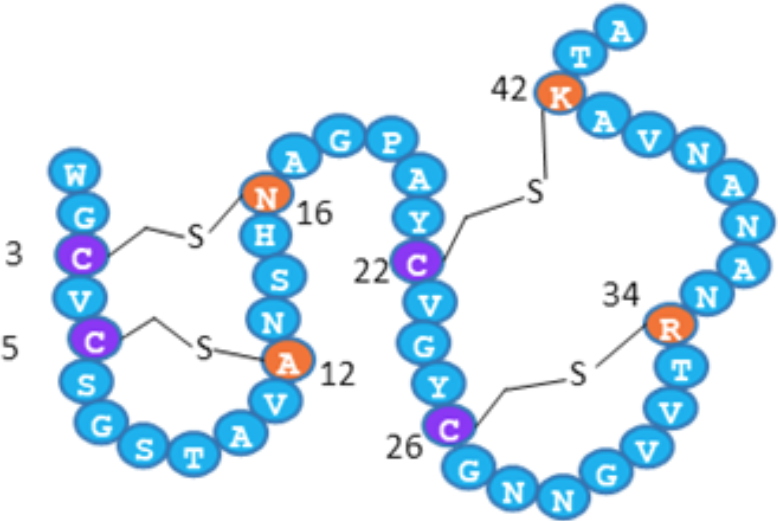
Figure 4: Double hairpin-like structure of RumC1. Cysteine residues bridged
via thioether bonds are shown in purple and their amino-acid partners are indicated in orange.
The heterologous maturation involving the specific radical-SAM enzyme RumMc1 establishes a novel four thioether-bond containing network, which creates a previously undescribed double-hairpin folding (Figure 4). This original structure confers to RumC1 a remarkable bactericidal activity against a wide range of target bacteria. Indeed, RumC1 is active at concentrations less than or equal to the micro-molar on laboratory strains or clinical isolates of Gram-positive pathogens, such as
Clostridium difficile,
Clostridium perfringens,
Enterococcus faecium and
Streptococcus pneumoniae, also including MDR strains (Table 1).
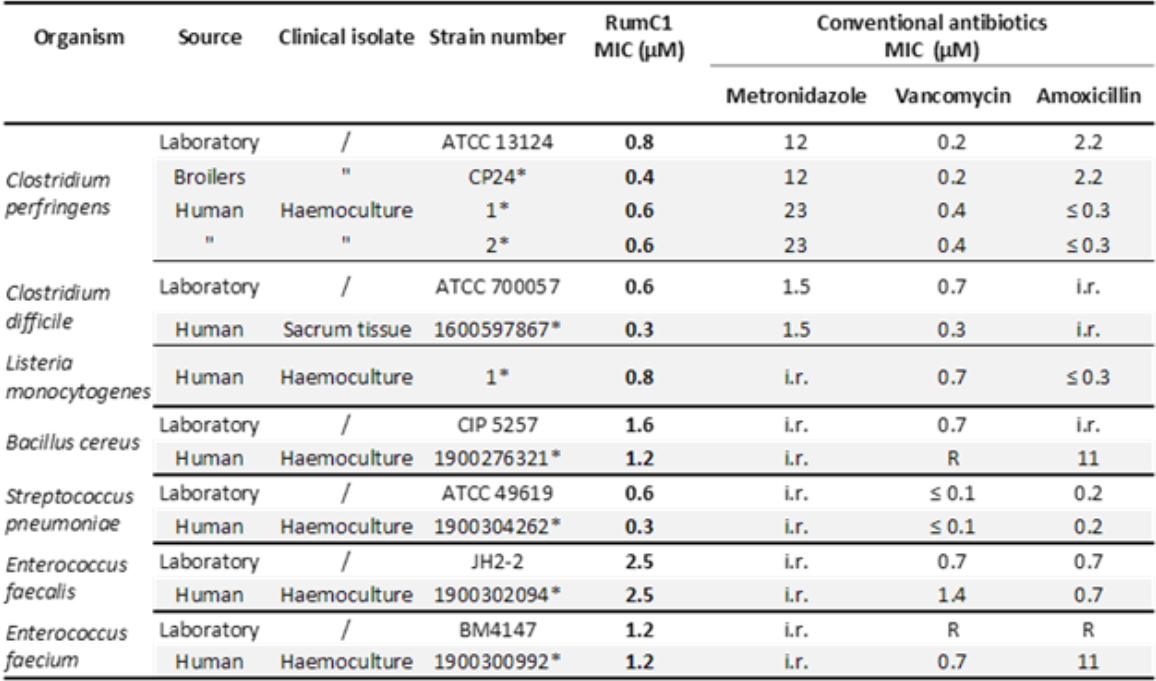
Table 1: Activity spectrum of RumC1 against laboratory and clinical Gram-positive pathogens. Acquired resistance to antibiotics were determined following the EUCAST 2019 clinical breakpoint tables and are indicated by "R”, whereas “i.r.” refers to “intrinsic resistance”, and * to the laboratory isolate references.
Moreover, no resistant bacterial clones have emerged following prolonged exposure to RumC1. Added to this efficacy, RumC1 is not significantly toxic to a number of human cell lines or intestinal explants. Finally, with regard to the mode of action of RumC1, we have shown that this compound is not "pore-forming" and that it seems to exert its bactericidal activity on
C. perfringens by blocking the synthesis of macromolecules (
i.e. nucleic acids, proteins and peptidoglycans), by most likely a yet undescribed mode of action.
 Publication Chiumento S, Roblin C, Kieffer-Jaquinod S, Tachon S, Leprètre C, Basset C, Aditiyarini D, Olleik H, Nicoletti C, Bornet O, Iranzo O, Maresca M, Hardré R, Fons M, Giardina T, Devillard E, Guerlesquin F, Couté Y, Atta M, Perrier J, Lafond M and Duarte V
Publication Chiumento S, Roblin C, Kieffer-Jaquinod S, Tachon S, Leprètre C, Basset C, Aditiyarini D, Olleik H, Nicoletti C, Bornet O, Iranzo O, Maresca M, Hardré R, Fons M, Giardina T, Devillard E, Guerlesquin F, Couté Y, Atta M, Perrier J, Lafond M and Duarte V Ruminococcin C, a promising antibiotic produced by a human gut symbiont.
Science Advances, 2019,
5(9): eaaw9969
 Funding
Funding RUMBA project ANR-15-CE21-0020
 Collaborations
Collaborations M. Lafond, Institut des Sciences Moléculaires de Marseille; F. Guerlesquin, Institut de Microbiologie de la Méditerranée, Marseille; E. Devillard, ADISSEO SAS France, Commentry; S. Torelli, Laboratoire Chimie et Biologie des Métaux, Grenoble; P. Polard, Lab. de Microbiologie et Génétique Moléculaires, Université Paul Sabatier, Toulouse.