 Nanoscale materials based on polypeptides are the objects of extensive researches for several years now. The self-assembling properties of these biopolymers make them particularly promising for the bottom-up design of materials, since they can undergo spontaneous organization into structures on nano- to macroscopic scales. Only weak forces, such as hydrogen and ionic bonds or hydrophobic and van der Waals interactions, are occurring within and between the polypeptides. However, their cumulative effect within well-ordered structures can result in highly stable objects. A variety of self-assembling nanoscale materials have been successfully designed with small peptides or proteins, and they have potential uses as scaffolds for three dimensional cell cultures (hydrogels), as carrier for drug delivery (nanotubes for instance), as structural template for enantiomeric separation or as peptide ink.
Nanoscale materials based on polypeptides are the objects of extensive researches for several years now. The self-assembling properties of these biopolymers make them particularly promising for the bottom-up design of materials, since they can undergo spontaneous organization into structures on nano- to macroscopic scales. Only weak forces, such as hydrogen and ionic bonds or hydrophobic and van der Waals interactions, are occurring within and between the polypeptides. However, their cumulative effect within well-ordered structures can result in highly stable objects. A variety of self-assembling nanoscale materials have been successfully designed with small peptides or proteins, and they have potential uses as scaffolds for three dimensional cell cultures (hydrogels), as carrier for drug delivery (nanotubes for instance), as structural template for enantiomeric separation or as peptide ink.
 Proteins are envisaged as self-assembling component of nanomaterials. Their size and their stable three-dimensional structure make possible the design of a larger variety of self-assembling objects. Nowadays, however, the
de novo design of a structured protein is impossible. Understanding the link between the information encoded in the amino acid sequence and the three dimensional structure of a protein, as well as how this structure can be formed spontaneously remains one of the big challenges in biological science. For that reason the design of self-assembling nanostructured objects is based on existing proteins whose properties (such as the propensity to form oligomers according to a particular network) are created by protein engineering.
Proteins are envisaged as self-assembling component of nanomaterials. Their size and their stable three-dimensional structure make possible the design of a larger variety of self-assembling objects. Nowadays, however, the
de novo design of a structured protein is impossible. Understanding the link between the information encoded in the amino acid sequence and the three dimensional structure of a protein, as well as how this structure can be formed spontaneously remains one of the big challenges in biological science. For that reason the design of self-assembling nanostructured objects is based on existing proteins whose properties (such as the propensity to form oligomers according to a particular network) are created by protein engineering.
 Nano-structured materials made of milk proteins: One of our projects is the investigation of possibilities to form nano-structured objects and multiscale biomaterials from natural milk proteins. These proteins can be produced at industrial scales; they are used in the food industry, to manufacture woven and as components within cosmetics. The most abundant and the best known of these proteins are the caseins, α-lactalbumin and ß-lactoglobulin; they are used as models for studies on various aspects of protein physical-chemistry: protein stabilities and refolding, aggregation… Depending on the considered protein and on the physico-chemical conditions, a variety of nano-scaled objects can be obtained.
Nano-structured materials made of milk proteins: One of our projects is the investigation of possibilities to form nano-structured objects and multiscale biomaterials from natural milk proteins. These proteins can be produced at industrial scales; they are used in the food industry, to manufacture woven and as components within cosmetics. The most abundant and the best known of these proteins are the caseins, α-lactalbumin and ß-lactoglobulin; they are used as models for studies on various aspects of protein physical-chemistry: protein stabilities and refolding, aggregation… Depending on the considered protein and on the physico-chemical conditions, a variety of nano-scaled objects can be obtained.
 Within the milk, caseins form micelles of diameters around 300 nm containing a nano-cluster of amorphous calcium phosphate, while α-lactalbumin and ß-lactoglobulin are soluble proteins.
In vitro and depending on the conditions, these proteins can self-assemble into various nano-structured objects. The diversity of possibilities is illustrated with α-lactalbumin in
Figure 3. Upon incubation at high temperature and in the presence of lysozyme, a hen egg white protein, α-lactalbumin forms droplet-like supramolecular structures on micrometer scale, the so-called coacervates (Figure 3; top/right). These objects are porous and show substructures on the nanometer scale. Upon incubation at acidic pH, amyloid fibrils are obtained (Figure 3; top/left). Their width is around 20 nm and their length up to several hundreds of nanometers. Tubular assemblies are obtained upon treatment with a protease and incubation at high temperature (Figure 3; bottom/left). In contrast to amyloid fibrils, these nanotubes require the presence of calcium ions for their formation, and a regular pattern can be detected on their surface.
Within the milk, caseins form micelles of diameters around 300 nm containing a nano-cluster of amorphous calcium phosphate, while α-lactalbumin and ß-lactoglobulin are soluble proteins.
In vitro and depending on the conditions, these proteins can self-assemble into various nano-structured objects. The diversity of possibilities is illustrated with α-lactalbumin in
Figure 3. Upon incubation at high temperature and in the presence of lysozyme, a hen egg white protein, α-lactalbumin forms droplet-like supramolecular structures on micrometer scale, the so-called coacervates (Figure 3; top/right). These objects are porous and show substructures on the nanometer scale. Upon incubation at acidic pH, amyloid fibrils are obtained (Figure 3; top/left). Their width is around 20 nm and their length up to several hundreds of nanometers. Tubular assemblies are obtained upon treatment with a protease and incubation at high temperature (Figure 3; bottom/left). In contrast to amyloid fibrils, these nanotubes require the presence of calcium ions for their formation, and a regular pattern can be detected on their surface.
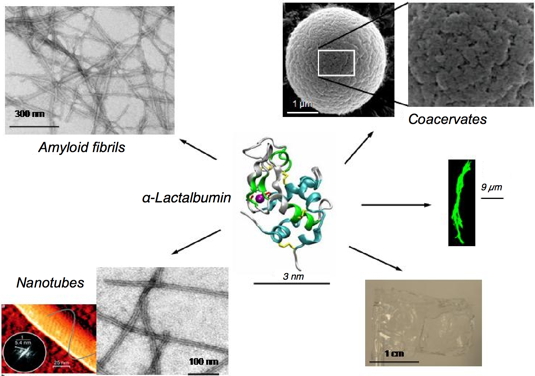
Figure 3: Various materials obtained by self-assembling α-lactalbumin.
Within the structure of the protein, the purple ball represents a tightly bound calcium ion. Coacervates are obtained at neutral pH in the absence of calcium but in the presence of lysozyme (Nigen et al.
Biochemistry, 2007). Amyloid fibrils are obtained at acidic pH. Nanotubes are built of peptides released from α-lactalbumin limited-proteolysis (Graveland-Bikker et al.,
NanoLetters, 2006). Fibres on the micrometer scale can be obtained by addition of anionic lipids vesicles (green fibres; adapted from Zhao et al.,
Biochemistry, 2004). Large scale film can be casted from
α-lactalbumin solubilized in diverse alcohols.
 These nano-objects can further assemble and form structures at larger scales (Figure 3). Upon incubation in presence of anionic lipid vesicles, larger fibres are obtained. Their width is around 1 µm and their length several tens of micrometers. When the solvent is an alcohol, the fibrils can assemble and form large scale films (Figure 3). Within water, the fibrils can form thixotropic hydrogels; several applications for these hydrogels are investigated.
These nano-objects can further assemble and form structures at larger scales (Figure 3). Upon incubation in presence of anionic lipid vesicles, larger fibres are obtained. Their width is around 1 µm and their length several tens of micrometers. When the solvent is an alcohol, the fibrils can assemble and form large scale films (Figure 3). Within water, the fibrils can form thixotropic hydrogels; several applications for these hydrogels are investigated.
 Understanding the driving forces governing protein assembly requires the characterization of interactions at molecular level (Delphine Salvadore
thesis and Salvatore
et al.,
Biomacromolecules, 2011a,
Biomacromolecules, 2011b). We focus on two homologous oppositely charged proteins: lysozyme (LYS) and α-lactalbumin (aLAC), which can assemble into microspheres. The assembly early steps were characterized through the identification of interacting surfaces monitored at residue level by NMR chemical shift perturbations, by titrating one
15N-labelled protein with its unlabelled partner. While α-lactalbumin has a narrow interacting site, lysozyme has interacting sites scattered on a broad surface. The further assembly of these rather unspecific heterodimers into tetramers leads to the establishment of well defined interaction sites. Within the tetramers, most of the electrostatic charge patches on the protein surfaces are shielded. Then, hydrophobic interactions, which are possible because α-lactalbumin is in a partially folded state, become preponderant leading to the formation of larger oligomers (Figure 4). This approach will be particularly useful for rationalizing the design of protein assemblies as nanoscale devices.
Understanding the driving forces governing protein assembly requires the characterization of interactions at molecular level (Delphine Salvadore
thesis and Salvatore
et al.,
Biomacromolecules, 2011a,
Biomacromolecules, 2011b). We focus on two homologous oppositely charged proteins: lysozyme (LYS) and α-lactalbumin (aLAC), which can assemble into microspheres. The assembly early steps were characterized through the identification of interacting surfaces monitored at residue level by NMR chemical shift perturbations, by titrating one
15N-labelled protein with its unlabelled partner. While α-lactalbumin has a narrow interacting site, lysozyme has interacting sites scattered on a broad surface. The further assembly of these rather unspecific heterodimers into tetramers leads to the establishment of well defined interaction sites. Within the tetramers, most of the electrostatic charge patches on the protein surfaces are shielded. Then, hydrophobic interactions, which are possible because α-lactalbumin is in a partially folded state, become preponderant leading to the formation of larger oligomers (Figure 4). This approach will be particularly useful for rationalizing the design of protein assemblies as nanoscale devices.
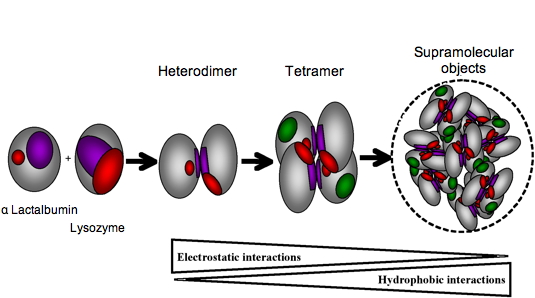
Figure 4: Schematic representation of aLAC-LYS dimer and tetramer formation. The LYS interacting surface determined experimentally is represented in purple and the residues interacting in a cooperative manner are represented in red. Residues of aLAC involved in dimer formation are supposed to be located in a similar region of that determined experimentally for hLAC (purple) and additional binding residues correspond to those near the calcium binding site (red). The aLAC is slightly larger than LYS since the molten globule state of LAC is known to be less compact that the native state. A first binding event involves purple patches whereas the second binding event for tetramer formation involves red patches. Oligomerisation into supramolecular objects is mainly driving by hydrophobic associations favoured by the exposition of hydrophobic patches (green) on the surface of molten globule of aLAC.
Protein-based conductive nanowires: The project we propose is based on the prion-domain of Het-s, a protein of a filamentous fungus (Podospora anserina). Within the Het-s protein, the prion domain is associated to a globular domain, which carries the recognition function. The prion-domain forms amyloid fibers independently of the globular domain (either it is present or not), and this is not related to a dysfunction, but it is necessary to the function of the whole protein.
We have chosen this prion-domain for our project because its structure has been determined at atomic resolution by solid-state nuclear magnetic resonance (NMR) (Wasmer
et al.,
Science, 2008). The structure of the fiber is a solenoid with a triangular hydrophobic core.
 For the design of our first generation of nanowires (Christophe Horvath
thesis), we have replaced the natural globular domain of Het-s by a redox protein able to transfer electrons. The first protein we have used is the rubredoxin containing a FeIII bound to four S-H groups of the protein (from four cysteins), this is the simplest version of the so-called FeS clusters. We have chosen this protein because of its robustness and easy handling. The principle is described in
Figure 5. Genes coding for the two proteins were associated within a single plasmid, which was internalized into bacteria (E. coli). Upon induction, bacteria over-expressed the resulting chimeric protein, which could be purified by chromatography. Upon purification the chimeric protein can be kept monomeric in urea; the formation of fiber by the prion domain can be induced by dialysis. At the end of the process, we obtain fibers decorated with redox domains which are, hopefully, able to transfer electrons by hopping (Figure 5).
For the design of our first generation of nanowires (Christophe Horvath
thesis), we have replaced the natural globular domain of Het-s by a redox protein able to transfer electrons. The first protein we have used is the rubredoxin containing a FeIII bound to four S-H groups of the protein (from four cysteins), this is the simplest version of the so-called FeS clusters. We have chosen this protein because of its robustness and easy handling. The principle is described in
Figure 5. Genes coding for the two proteins were associated within a single plasmid, which was internalized into bacteria (E. coli). Upon induction, bacteria over-expressed the resulting chimeric protein, which could be purified by chromatography. Upon purification the chimeric protein can be kept monomeric in urea; the formation of fiber by the prion domain can be induced by dialysis. At the end of the process, we obtain fibers decorated with redox domains which are, hopefully, able to transfer electrons by hopping (Figure 5).
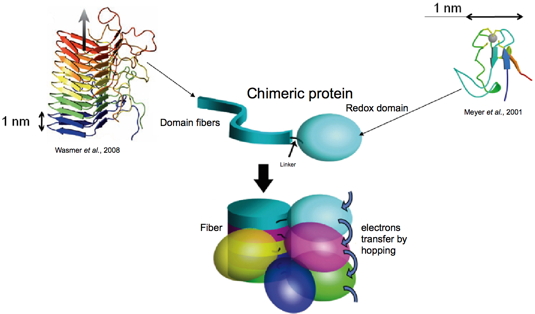
Figure 5: Strategy for the design of protein-based conductive nanowires.