 Reactivity of zinc fingers: Oxidative stress and metallic stress
Reactivity of zinc fingers: Oxidative stress and metallic stress
We are interested in understanding the properties of metal sites in proteins and zinc fingers, especially. Zinc fingers are small protein domains where a Zn
2+ ion is bound to four cysteine or histidine residues in a tetrahedral geometry, corresponding to the general formula Zn(Cys)
4-x(His)
x (x=0, 1 or 2). Zinc finger proteins are ubiquitous in biology, representing ca. 8% of the human proteome, for instance. Most zinc finger sites have a structural role but they can be reactive toward electrophiles and some proteins use this property for catalysis purpose. Examples of proteins with a reactive zinc finger site include the DNA repair protein ADA and farnesyl transferases. They catalyze the transfer of alkyl groups on cysteine sulfur. More recently, zinc finger sites were proposed to act as redox switch for the detection and signaling of reactive oxygen species. In order to understand the properties of zinc finger sites but also of other metal sites, we use models made of short peptides. These metallopeptides allow in depth studies at the molecular level that are not possible with entire proteins.

 Design of new zinc finger models of various folds
Design of new zinc finger models of various folds
Zinc finger domains in proteins are generally made of more than 40-70 amino acids. Most zinc fingers can be classified into a limited number of fold families with the treble clef, the zinc ribbon, the classical ßßα and the zinc necklace fold being the most important families. Except for classical ßßα zinc fingers, short linear peptides often fail in folding properly around the zinc ion and reproducing these folds. We have created a peptide design based on cyclic peptides with a linear tail (CPLT) that allows the successful structural modeling of zinc finger of various folds (treble clef, zinc ribbon, loosened zinc ribbon) with peptides made of only ca. 20 amino acids (
Figure 1). These peptidic models are very stable and robust: they resist to conditions in which proteins may not be stable (high pH, concentration or temperature). In addition, we have shown that these structurally relevant models reproduce the reactivity of the native zinc finger proteins that they mimic.
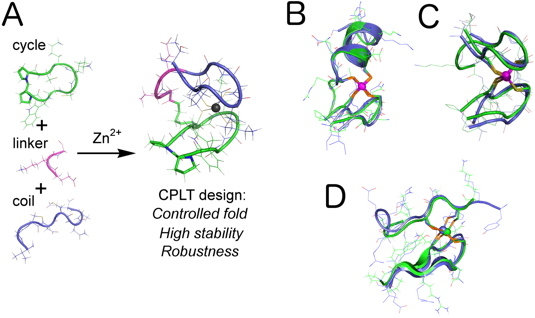 Figure 1: (A) Summary of the CPLT design for zinc fingers. (B, C and D) Examples of 1H NMR solution structures of CPLT-based models (green) superimposed to the crystallographic structures of various zinc fingers (blue, B = treble clef, C = zinc ribbon and D = loosened zinc ribbon).
REFERENCES
Sénèque O, Bourles E, Lebrun V, Bonnet E, Dumy P and Latour JM. Cyclic peptides bearing a side-chain tail: A tool to model the structure and reactivity of protein zinc sites. Angewandte Chemie International Edition, 2008
Figure 1: (A) Summary of the CPLT design for zinc fingers. (B, C and D) Examples of 1H NMR solution structures of CPLT-based models (green) superimposed to the crystallographic structures of various zinc fingers (blue, B = treble clef, C = zinc ribbon and D = loosened zinc ribbon).
REFERENCES
Sénèque O, Bourles E, Lebrun V, Bonnet E, Dumy P and Latour JM. Cyclic peptides bearing a side-chain tail: A tool to model the structure and reactivity of protein zinc sites. Angewandte Chemie International Edition, 2008
Sénèque O, Bonnet E, Joumas FL and Latour JM. Cooperative metal binding and helical folding in model peptides of treble-clef zinc fingers. Chemistry-A European Journal, 2009
Jacques A, Mettra B, Lebrun V, Latour JM and Sénèque O. On the design of zinc-finger models with cyclic peptides bearing a linear tail. Chemistry–A European Journal, 2013

 Reactivity of zinc fingers toward reactive oxygen species
Reactivity of zinc fingers toward reactive oxygen species
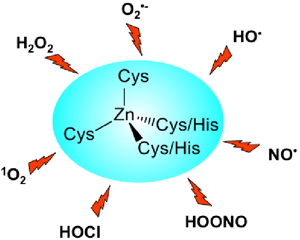 | |
Figure 2: Reactive oxygen and nitrogen species likely to damage zinc fingers in vivo. | |
Several proteins involved in the defense against oxidative stress such as Hsp33, RsrA or ChrR have been proposed to sense reactive oxygen species thanks to a zinc finger site that acts as a redox switch. We are currently studying the reactivity of zinc finger models toward various reactive oxygen and nitrogen species ( Figure 2). |
 Hydrogen peroxide H2O2
Hydrogen peroxide H2O2
|  |
|
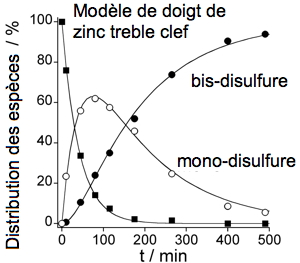
Figure 3: Kinetic study of the reaction of a CPLT-based model of treble clef zinc finger with H2O2. |
| |
In order to get mechanistical insights into this reactivity, we identify the products of the reaction and possible intermediates by means of HPLC purification and ESI/MS analyses and we measure the kinetics by several techniques (
Figure 3). First we have focused on H
2O
2. Our results show that the oxidation of zinc finger by H
2O
2 is very slow with half-lifes of zinc fingers ranging from hours to days at H
2O
2 concentrations corresponding to a severe stress. This mean that zinc fingers are not likely targets of H
2O
2 and cannot be used as H
2O
2 sensors by cells. However, he study of the reactivity of zinc fingers with H
2O
2 has revealed that the nucleophilic reactivity of zinc finger is mainly dictated by the number of coordinated cysteines but is highly modulated by the conformational flexibility of the zinc finger: the more flexible the zinc finger, the higher the reactivity.
REFERENCES
Bourles E, Isaac M, Lebrun C, Latour JM and Sénèque O. Oxidation of Zn(Cys)4 zinc finger peptides by O2 and H2O2: Products, mechanism and kinetics. Chemistry–A European Journal, 2011
Isaac M, Latour JM and Sénèque O. Nucleophilic reactivity of Zinc-bound thiolates: Subtle interplay between coordination set and conformational flexibility. Chemical Science, 2012
 Hypochlorous acid HOCl
Hypochlorous acid HOCl
In mammals, hypochlorous acid (HOCl) is produced in macrophages to destroy bacteria. As a first line in the defense against HOCl, some bacteria use proteins that are capable of sensing HOCl, such as Hsp33. Although it has been demonstrated by the group of Ursula Jakob that Hsp33 senses HOCl through the oxidation of its Zn(Cys)
4 site, no kinetic data were reported for such a reaction. We have used a CPLT model of the zinc finger site of Hp33 to measure the rate of the reaction of this site with HOCl: it is very fast with a rate constant approaching the diffusion limit. The zinc finger site of Hsp33 reacts with HOCl faster than methionine or cysteine, which are the amino acids that react the fastest with HOCl. This supports the role of Hsp33 in HOCl sensing in bacteria.
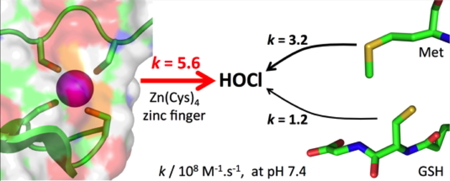 Figure 4: Reactivity toward HOCl: Zn(Cys)4 zinc finger vs methionine or glutathione.
REFERENCE
Lebrun V, Ravanat JL, Latour JM and Sénèque O. Near diffusion-controlled reaction of a Zn(Cys)4 zinc finger with hypochlorous acid. Chemical Science, 2016
Figure 4: Reactivity toward HOCl: Zn(Cys)4 zinc finger vs methionine or glutathione.
REFERENCE
Lebrun V, Ravanat JL, Latour JM and Sénèque O. Near diffusion-controlled reaction of a Zn(Cys)4 zinc finger with hypochlorous acid. Chemical Science, 2016
 Singlet oxygen 1O2
Singlet oxygen 1O2
Singlet oxygen (
1O
2) is another powerful oxidant produced in macrophages. We have shown the reaction between
1O
2 and our CPLT model of Zn(Cys)
4 treble-clef zinc fingers is very efficient. It leads to the oxidation of zinc-bound cysteinates into sulfinates (RSOO–), which destabilizes the zinc site and favors the release of zinc from the peptide as well as the unfolding of the peptide in physiological conditions. Hence,
1O
2 is able to destroy Zn(Cys)
4 sites.We have also demonstrated that the Zn(Cys)
2His)
2 site of a classical zinc finger is far less reactive in similar conditions.
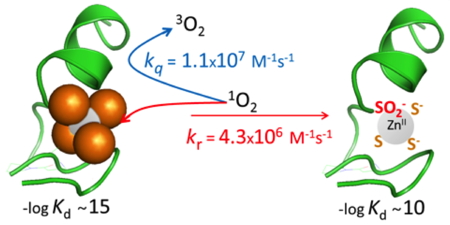 Figure 5: Reaction between a treble-clef Zn(Cys)4 zinc finger and singlet oxygen.
REFERENCES
Lebrun V, Tron A, Scarpantonio L, Lebrun C, Ravanat JL, Latour JM, McClenaghan ND and Sénèque O. Efficient oxidation and destabilization of Zn(Cys)4 zinc fingers by singlet oxygen. Angewandte Chemie-International Edition, 2014
Figure 5: Reaction between a treble-clef Zn(Cys)4 zinc finger and singlet oxygen.
REFERENCES
Lebrun V, Tron A, Scarpantonio L, Lebrun C, Ravanat JL, Latour JM, McClenaghan ND and Sénèque O. Efficient oxidation and destabilization of Zn(Cys)4 zinc fingers by singlet oxygen. Angewandte Chemie-International Edition, 2014
Lebrun V, Tron A, Lebrun C, Latour JM, McClenaghan ND and Sénèque O. Reactivity of a Zn(Cys)2 (His)2 zinc finger with singlet oxygen: Oxidation directed toward cysteines but not histidines. Chemistry - A European Journal, 2015
Chabert V, Lebrun V, Lebrun C, Latour JM and Sénèque O. Model peptide for anti-sigma factor domain HHCC zinc fingers: High reactivity toward 1O2 leads to domain unfolding. Chemical Science, 2019

 Reactivity of zinc fingers toward metal ions or metal complexes
Reactivity of zinc fingers toward metal ions or metal complexes
Gold salts have a high affinity for thiols and are thus likely to target any protein containing cysteines. Gold(I) salts have been used for a long time in the treatment of several diseases such as rheumatoid arthritis and gold(III) salts have promising antitumoral activities. However, their mechanism of action is still unknown. Among possible hypothesis, they could interact with some zinc finger proteins and we have studied this with our models. The reaction of the gold(III) complex Au
IIIterpy (terpy = terpyridine) with a CPLT (ZnL
ZR) modeling the Zn(Cys)
4 zinc ribbon zinc finger is very fast (k ≈ 10
5 M
-1 s
-1). The reaction product is a peptide with two cysteines oxidized as a disulfide and the two others bound to a gold(I) ion in a S–Au
I–S motif. This shows that a gold(III) complex can target and destroy a Zn(Cys)
4 zinc finger site. In the presence of an excess of thiols mimicking cellular glutathione, the reaction between Au
IIIterpy and ZnL
ZR leads to the removal of zinc from the peptide and the formation of the Au
2L
ZR complex with two S–Au
I–S motifs. This shows that the destruction of Zn(Cys)
4 zinc finger sites by gold complexes is possible in an environment close to cellular one.
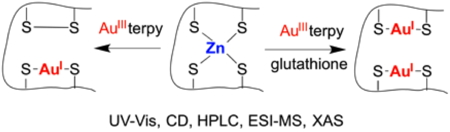 Figure 6: Reactivity of a Zinc Ribbon Zn(Cys)4 site and a gold(III) complex.
REFERENCE
Jacques A, Lebrun C, Casini A, Kieffer I, Proux O, Latour JM and Sénèque O. Reactivity of Cys4 Zinc finger domains with Gold(III) complexes: Insights into the formation of "gold fingers". Inorganic Chemistry, 2015
Figure 6: Reactivity of a Zinc Ribbon Zn(Cys)4 site and a gold(III) complex.
REFERENCE
Jacques A, Lebrun C, Casini A, Kieffer I, Proux O, Latour JM and Sénèque O. Reactivity of Cys4 Zinc finger domains with Gold(III) complexes: Insights into the formation of "gold fingers". Inorganic Chemistry, 2015
 Iron proteins models
Iron proteins models
Our CPLT model peptide designed for the modeling of zinc ribbon zinc finger (L
ZR) is able to form tight and stable complexes with Fe
2+ and Fe
3+ in water around pH 7. It reproduces all the spectroscopic feature of reduced and oxidized rubredoxin, an electron transfer protein that bears a Fe(Cys)
4 site. FeL
ZR has a redox potential of +144 mV / NHE and is able to cycle between the Fe
2+ and Fe
3+ states like rubredoxin (
Figure 7). It constitutes the most stable model complex of rubredoxin so far with such a limited number of amino acids. The properties of this complex demonstrate that redox functionality may be implemented in CPLT-based metallopeptides due to their robustness.
 Figure 7: (A) UV-Vis and (B) CD spectra of FeIILZR and FeIIILZR. (C) Redox cycling of FeLZR monitored following the absorbance at 491 nm.
REFERENCES
Jacques A, Clémancey M, Blondin G, Fourmond V, Latour JM and Sénèque O. A cyclic peptide-based redox-active model of rubredoxin. Chemical Communications (Camb), 2013
Figure 7: (A) UV-Vis and (B) CD spectra of FeIILZR and FeIIILZR. (C) Redox cycling of FeLZR monitored following the absorbance at 491 nm.
REFERENCES
Jacques A, Clémancey M, Blondin G, Fourmond V, Latour JM and Sénèque O. A cyclic peptide-based redox-active model of rubredoxin. Chemical Communications (Camb), 2013
Jacques A, Latour JM and Sénèque O. Peptide-based FeS4 complexes: The zinc ribbon fold is unsurpassed to stabilize both the FeII and FeIII states. Dalton Transactions, 2014
 Smart luminescent probes and contrast agents
Smart luminescent probes and contrast agents
Capitalizing on our knowledge concerning the design, the synthesis and the coordination properties of zinc fingers, we are developing smart peptide-based probes for the sensing of metal cations and biomolecules in biological media. These probes are inspired by binding sites of proteins (metal binding sites or DNA/RNA binding domains for instance) to benefit from their unique recognition properties and they make use of lanthanide ions for their luminescence or magnetic properties.

 Luminescent probes for the detection of Zn2+
Luminescent probes for the detection of Zn2+
In order to elaborate zinc-selective probes, we have chosen the classical ββα zinc finger as a scaffold. This type of protein binds Zn
2+ with a high affinity and a high selectivity with respect to other physiological cations. By introducing a lanthanide complex and a suitable sensitizing antenna at well-chosen positions in the zinc finger sequence, we have obtained zinc selective probes that can detect and signal Zn
2+ with emission wavelength ranging from the visible to the NIR.
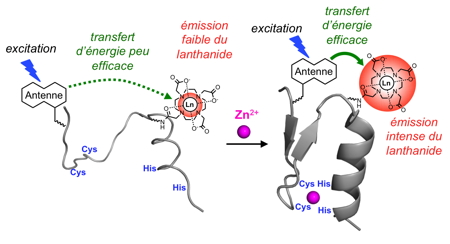 Figure 8: Principle of Zn2+ sensing by a luminescent zinc finger.
REFERENCES
Cepeda C, Raibaut L, Fremy G, Eliseeva SV, Romieu Z, Pecaut J, Boturyn D, Petoud S and Sénèque O. Using native chemical ligation for site-specific synthesis of hetero-bis-lanthanide peptide conjugates: Application to ratiometric visible or near-infrared detection of Zn2+, Chemistry A European Journal, 2020
Figure 8: Principle of Zn2+ sensing by a luminescent zinc finger.
REFERENCES
Cepeda C, Raibaut L, Fremy G, Eliseeva SV, Romieu Z, Pecaut J, Boturyn D, Petoud S and Sénèque O. Using native chemical ligation for site-specific synthesis of hetero-bis-lanthanide peptide conjugates: Application to ratiometric visible or near-infrared detection of Zn2+, Chemistry A European Journal, 2020
Fremy G, Raibaut L, Cepeda C, Sanson M, Boujut M and Sénèque O. A novel DOTA-like building block with a picolinate arm for the synthesis of lanthanide complex-peptide conjugates with improved luminescence properties, Journal of Inorganic Biochemistry, 2020
Isaac M, Pallier A, Szeremeta F, Bayle PA, Barantin L, Bonnet CS and Sénèque O.
MRI and luminescence detection of Zn2+ with a lanthanide complex-zinc finger peptide conjugate.
Chemical Communications (Camb), 2018
Isaac M, Raibaut L, Cepeda C, Roux A, Boturyn D, Eliseeva SV, Petoud S and Sénèque O. Luminescent zinc fingers: Zn-responsive neodymium near-infrared emission in water. Chemistry-a European Journal, 2017

 Luminescent probes for the detection of Cu+
Luminescent probes for the detection of Cu+
We have also designed a probe for Cu
+ inspired by the protein CusF, which binds selectively this ion thanks to two methionines, one histidine and one tryptophan. The cyclic probes associate a terbium (Tb
3+) complex to the CusF binding loop and it is able to establish a cation/π interaction between the tryptophan side chain and the Cu
+ ion, as does the protein CusF. This interaction triggers the enhancement of the terbium emission.
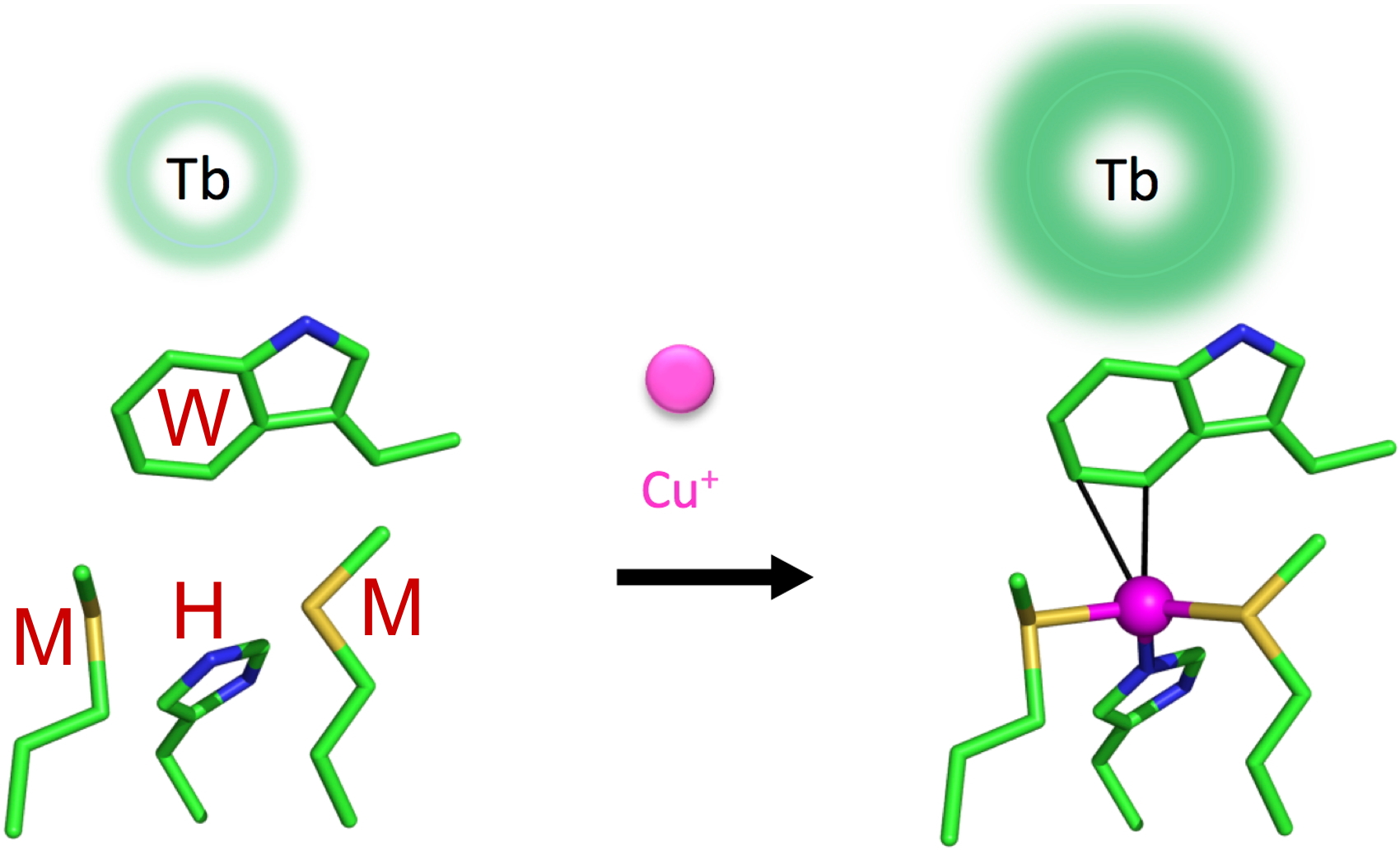 Figure 9: Bio-inpired luminescent probe for the detection of Cu+.
REFERENCES
Isaac M, Denisov SA, McClenaghan ND and Sénèque O. Bioinspired luminescent Europium-based probe capable of discrimination between Ag+ and Cu+. Inorganic Chemistry, 2021
Figure 9: Bio-inpired luminescent probe for the detection of Cu+.
REFERENCES
Isaac M, Denisov SA, McClenaghan ND and Sénèque O. Bioinspired luminescent Europium-based probe capable of discrimination between Ag+ and Cu+. Inorganic Chemistry, 2021
Cepeda C, Denisov SA, Boturyn D, McClenaghan ND and Sénèque O. Ratiometric luminescence detection of Copper(I) by a resonant system comprising two antenna/lanthanide pairs. Inorganic Chemistry, 2021
Roux A, Isaac M, Chabert V, Denisov SA, McClenaghan ND and Seneque O. Influence of amino acid sequence in a peptidic Cu(+)-responsive luminescent probe inspired by the copper chaperone CusF. Organic and Biomolecular Chemistry, 2018
Isaac M, Denisov SA, Roux A, Imbert D, Jonusauskas G, McClenaghan ND and Sénèque O. Lanthanide luminescence modulation by cation-pi interaction in a bioinspired scaffold: Selective detection of copper(I). Angewandte Chemie - International Edition, 2015

 Selective detection of the AU-rich element of messenger RNAs
Selective detection of the AU-rich element of messenger RNAs
We have designed of a luminescent sensor based upon the zinc finger protein TIS11d, that allows for the selective time-resolved detection of the UUAUUUAUU sequence of 3’-untranslated region of messenger RNA. This sensor, which was obtained by chemical synthesis using native chemical ligation, is composed of the tandem ZF RNA binding domain of TIS11d functionalized with a luminescent Tb
3+ complex on one of the ZFs and a sensitizing antenna on the other. It provides the proof of principle that an RNA binding protein can be re-engineered as an RNA sensor and, more generally, that tunable synthetic luminescent probes for biomolecules can be obtained by modifying biomolecule-binding domains of proteins.
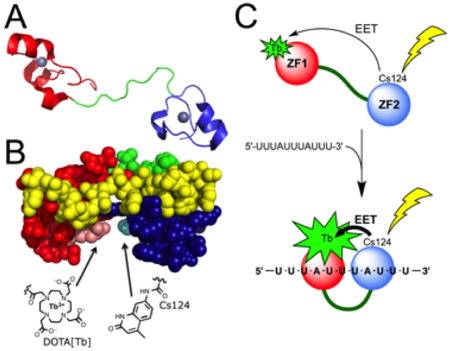 Figure 10: Principle of mRNA detection by a synthetic protein-based luminescent sensor.
REFERENCE
Raibaut L, Vasseur W, Shimberg GD, Saint-Pierre C, Ravanat JL, Michel SLJ and Sénèque O. Design of a synthetic luminescent probe from a biomolecule binding domain: Selective detection of AU-rich mRNA sequences. Chemical Science, 2017
Figure 10: Principle of mRNA detection by a synthetic protein-based luminescent sensor.
REFERENCE
Raibaut L, Vasseur W, Shimberg GD, Saint-Pierre C, Ravanat JL, Michel SLJ and Sénèque O. Design of a synthetic luminescent probe from a biomolecule binding domain: Selective detection of AU-rich mRNA sequences. Chemical Science, 2017